Cuckoo filter
Reference: Fan, Andersen, Kaminsky & Mitzenmacher: "Cuckoo Filter: Practically Better Than Bloom"
See also the page: Cuckoo versus bloom filters
What it is
A cuckoo filter is conceptually similar to a bloom filter, even though the underlying algorithm is quite different.
The cuckoo filter is similar to a Set. Hashable objects can be pushed into the filter, but objects cannot be extracted from the filter. Querying the filter, i.e. asking whether an object is in a filter is fast, but cuckoo filters has a certain probability of falsely returning true when querying about an object not actually in the filter. When querying an object that is in the filter, it is guaranteed to return true.
A cuckoo filter is defined by two parameters, F and its length L. Memory usage is F*L/2 bytes plus 50-ish bytes of overhead, and the false positive rate is approximately 9*(N/L)*(2^-F) where N is the number of elements in the filter.
Querying (in)
Querying takes one cache access plus either 1 or 2 random memory access depending on the values of N, F and L, and so can be thought of as being constant. When querying about an object in the filter, it is guaranteed to return true, unless deletion operations have been done on the filrter. Querying about objects not in the filter returns true with a probability approximately to 9*(N/L)*(2^-F).
In general, two distinct objects A and B will have a 1/(2^F-1) chance of sharing so-called "fingerprints". Independently, each object is assigned two "buckets" in the range 1:L/4, and inserted in an arbitrary of the two buckets. If objects A and B share fingerprints, and object A is in one of B's buckets, the existence of A will make a query for B return true, even when it's not in the filter.
Pushing (push!)
Only hashable objects can be inserted into the filter. Inserting time is stochastic, but its expected duration is proportional to 1/(1 - N/L). To avoid infinite insertion times as N approaches L, an insert operation may fail if the filter is too full. A failed push operation returns false.
Pushing may yield false positives: If an object A exists in the filter, and querying for object B would falsely return true, then pushing B to the filter has a probability 1/2 + 1/2 * N/L of returning true while doing nothing, because B is falsely believed to already be in the filter.
Deletion (pop!)
Objects can be deleted from the filter. Deleting operation also exhibits false positives: If B has been pushed to the filter, falsely returning success, then deleting A will also delete B.
Deletion of objects is fast and constant time, except if the filter is at full capacity. In that case, it will attempt to self-organize after a deletion to allow new objects to be pushed. This might take up to 200 microseconds.
FastCuckoo and SmallCuckoo
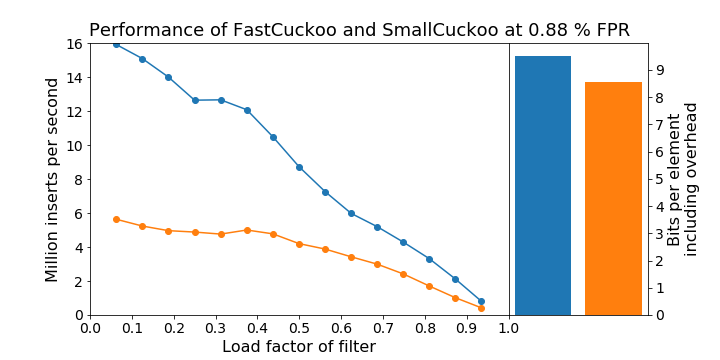
Probably.jl comes with two different implementations of the cuckoo filter: FastCuckoo and SmallCuckoo. The latter uses a more complicated encoding scheme to achieve a slightly smaller memory footprint, but which also make all operations slower. The following plot shows how the speed of pushing objects depend on the load factor, i.e. how full the filter is, and how FastCuckoos are ~2.5x faster than SmallCuckoos, but that the SmallCuckoo uses about 10% less memory. FastCuckoo is displayed in blue, SmallCuckoo in orange.
Usage example
For this example, let's say I have a stream of kmers that I want to count. Of course I use BioJulia, so these kmers are represented by a DNAKmer{31} object. I suspect my stream has up to 2 billion different kmers, so keeping a counting Dict would use up all my memory. However, most kmers are measurement errors that only appear once and that I do not need spend memory on keeping count of. So I keep track of which kmers I've seen using a Cuckoo filter. If I see a kmer more than once, I add it to the Dict.
params = constrain(SmallCuckoo, fpr=0.02, capacity=2_000_000_000)
if params.memory > 4e9 # I'm only comfortable using 4 GB of memory for this
error("Too little memory :(")
end
filter = SmallCuckoo{params.F}(params.nfingerprints)
counts = Dict{Kmer, UInt8}() # Don't need to count higher than 255
for kmer in each(DNAKmer{31}, fastq_parser)
if kmer in filter
# Only add kmers we've seen before
count = min(0xfe, get(counts, kmer, 0x01)) # No integer overflow
counts[kmer] = count + 0x01
else
push!(filter, kmer)
end
endInterface
Construction
A cuckoo filter can be constructed directly from the two parameters F and L, where L is the number of fingerprint slots in the fingerprint. Remember that L should be a power-of-two:
julia> FastCuckoo{12}(2^32) # F=12, L=2^32, about 6 GiB in size
However, typically, one wants to construct cuckoo filters under some kind of constrains: Perhaps I need to store at least 1.1 billion distinct elements, with a maximal false positive rate of 0.004. For this purpose, use the constrain function.
This function takes a type and two of three keyword arguments:
fpr: Maximal false positive ratememory: Maximal memory usagecapacity: Minimum number of distinct elements it can contain
It returns a NamedTuple with the parameters for an object of the specified type which fits the criteria:
julia> constrain(SmallCuckoo, fpr=0.004, capacity=1.1e9)
(F = 11, nfingerprints = 2147483648, fpr = 0.002196371220581028, memory = 2952790074, capacity = 2040109466)Having passed false positive rate and capacity, the function determined the smallest possible SmallCuckoo that fits these criteria. We can see from the fields F and nfingerprints that such a SmallCuckoo can be constructed with:
SmallCuckoo{11}(2147483648)
Furthermore, we can also see that those particular constrains were quite unlucky: The actual false positive rate of this filter will be ~0.0022 (at full capacity), and its actual capacity will be ~2 billion elements. It is not possible to create a smaller SmallCuckoo which fits the given criteria.
Central functions
Base.in — Methodin(item, filter::AbstractCuckooFilter)Check if an item is in the cuckoo filter. This can sometimes erroneously return true, but never erroneously returns false, unless a pop! operation has been performed on the filter.
Base.push! — Methodpush!(filter::AbstractCuckooFilter, items...)Insert one or more items into the cuckoo filter. Returns true if all inserts was successful and false otherwise.
Base.pop! — Methodpop!(filter::AbstractCuckooFilter, item)Delete an item from the cuckoo filter, returning the filter. Does not throw an error if the item does not exist. Has a risk of deleting other items if they collide with the target item in the filter.
Examples
julia> a = FastCuckoo{12}(2^4); push!(a, 1); push!(a, 868)
julia> pop!(a, 1); # Remove 1, this accidentally deletes 868 also
julia> isempty(a)
trueMisc functions
Cuckoo filters supports the following operations, which have no cuckoo-specific docstring because they behave as stated in the documentation in Base:
Base.copy!
Base.copy
Base.sizeof # This one includes the underlying arrayBase.isempty — Methodisempty(x::AbstractCuckooFilter)Test if the filter contains no elements. Guaranteed to be correct.
Examples
julia> a = FastCuckoo{12}(1<<12); isempty(a)
trueBase.empty! — Methodempty!(x::AbstractCuckooFilter)Removes all objects from the cuckoo filter, resetting it to initial state.
Examples
julia> a = FastCuckoo{12}(1<<12); push!(a, "Hello"); isempty(a)
false
julia> empty!(a); isempty(a)
trueBase.union! — Methodunion!(dest::HyperLogLog{P}, src::HyperLogLog{P})Overwrite dest with the same result as union(dest, src), returning dest.
Examples
julia> # length(c) ≥ length(b) is not guaranteed, but overwhelmingly likely
julia> c = union!(a, b); c === a && length(c) ≥ length(b)
trueunion!(dst::AbstractCuckooFilter{F}, src::AbstractCuckooFilter{F})Attempt to add all elements of source filter to destination filter. If destination runs out of space, abort the copying and return (destination, false). Else, return (destination, true). Both filters must have the same length and F value.
Base.union — Methodunion(x::HyperLogLog{P}, y::HyperLogLog{P})Create a new HLL identical to an HLL which has seen the union of the elements x and y has seen.
Examples
julia> # That c is longer than a or b is not guaranteed, but overwhelmingly likely
julia> c = union(a, b); length(c) ≥ length(a) && length(c) ≥ length(b)
trueunion(x::AbstractCuckooFilter{F}, y::AbstractCuckooFilter{F})Attempt to create a new cuckoo fitler with the same length and F value as x and y, and with the union of their elements. If the new array does not have enough space, returns (newfilter, false), else returns (newfilter, true). Both filters must have the same length and F value.
Probably.loadfactor — Methodloadfactor(x::AbstractCuckooFilter)Returns fraction of filled fingerprint slots, i.e. how full the filter is.
Examples
julia> a = FastCuckoo{12}(1<<12);
julia> for i in 1:1<<11 push!(i, a) end; loadfactor(a)
0.5Probably.fprof — Functionfprof(::Type{AbstractCuckooFilter{F}}) where {F}
fprof(x::AbstractCuckooFilter)Get the false positive rate for a fully filled AbstractCuckooFilter{F}. The FPR is proportional to the fullness (a.k.a load factor).
fprof(sketch::CountMinSketch)Estimate the probability of miscounting an element in the sketch.
Probably.capacityof — Functioncapacityof(filter::AbstractCuckooFilter)Estimate the number of distinct elements that can be pushed to the filter before adding more will fail. Since push failures are probabilistic, this is not accurate, but for filters with a capacity of thousands or more, this is rarely more than 1% off.
Probably.constrain — Methodconstrain(T<:AbstractCuckooFilter; fpr=nothing, mem=nothing, capacity=nothing)Given a subtype of AbstractCuckooFilter and two of three keyword arguments, as constrains, optimize the elided keyword argument. Returns a NamedTuple with (F, nfingerprints, fpr, memory, capacity), which applies to an instance of the optimized CuckooFilter.
Examples
julia> # FastCuckoo with FPR ≤ 0.001, and memory usage ≤ 250_000_000 bytes
julia> c = constrain(FastCuckoo, fpr=0.001, memory=250_000_000)
(F = 14, nfingerprints = 134217728, fpr = 0.0005492605216655955, memory = 234881081,
capacity = 127506842)
julia> x = FastCuckoo{c.F}(c.nfingerprints); # capacity optimized
julia> fprof(x), sizeof(x), capacityof(x) # not always exactly the estimate
(0.0005492605216655955, 234881081, 127506842)